Scientific Programs
in the Institute for Computational BiomedicineComputational Neuroscience
Aksay Lab
Research Summary | Publications
 A function of many neural systems is to accumulate information over time and remember the summed value as a pattern of neural activity. This process can often be described mathematically as temporal integration. Integration appears to play a key role in many sensori-motor transformations, short-term memory behaviors, and decision making problems. The aims of this laboratory are to 1) determine the molecular, cellular, and circuit properties that enable integration, and 2) determine how these properties are modified to improve integration.
A function of many neural systems is to accumulate information over time and remember the summed value as a pattern of neural activity. This process can often be described mathematically as temporal integration. Integration appears to play a key role in many sensori-motor transformations, short-term memory behaviors, and decision making problems. The aims of this laboratory are to 1) determine the molecular, cellular, and circuit properties that enable integration, and 2) determine how these properties are modified to improve integration.Nirenberg lab
Research Summary | Publications
 The theme of our research is to advance basic understanding of computational neuroscience, and, in parallel, use what we learn to address practical problems that improve quality of life. Our work is organized around three major goals: To understand the codes neurons use and the transformations they perform To use this understanding to build neuroprosthetics, brain machine interfaces, and robots To use our knowledge of the codes and transformations to address key questions in systems neuroscience. Specifically, we’re leveraging our knowledge of coding at the single cell level to advance our understanding at the population level, to understand how populations collectively solve problems (e.g., represent images, extract information).
The theme of our research is to advance basic understanding of computational neuroscience, and, in parallel, use what we learn to address practical problems that improve quality of life. Our work is organized around three major goals: To understand the codes neurons use and the transformations they perform To use this understanding to build neuroprosthetics, brain machine interfaces, and robots To use our knowledge of the codes and transformations to address key questions in systems neuroscience. Specifically, we’re leveraging our knowledge of coding at the single cell level to advance our understanding at the population level, to understand how populations collectively solve problems (e.g., represent images, extract information).Victor lab
Research Summary | Publications
 Overall, we are interested in the design principles of sensory processing, both in the sensory periphery and in the brain, and how these design principles are implemented in biological hardware. We seek to determine the aspects of sensory information that are represented, the features of the activity of individual neurons and neural populations that support these representations, and realistic models for how these representations are transformed. These questions are addressed in the visual system, via neurophysiologic studies at the single-cell and multineuronal level (with Keith Purpura) and via psychophysical studies (with Mary Conte).
Overall, we are interested in the design principles of sensory processing, both in the sensory periphery and in the brain, and how these design principles are implemented in biological hardware. We seek to determine the aspects of sensory information that are represented, the features of the activity of individual neurons and neural populations that support these representations, and realistic models for how these representations are transformed. These questions are addressed in the visual system, via neurophysiologic studies at the single-cell and multineuronal level (with Keith Purpura) and via psychophysical studies (with Mary Conte).Computational Integrative Physiology
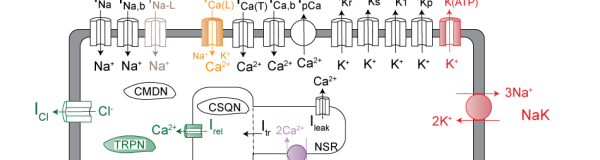
Christini lab
Research Summary | Publications
 Sudden cardiac death, primarily caused by ventricular arrhythmias, is a major public health problem – it is one of the leading causes of mortality, resulting in more than 350,000 annual deaths in the United States alone. Our group’s efforts are focused on improving our understanding of, and therapies for, cardiac arrhythmias. We primarily investigate biophysical mechanisms of electrophysiological instabilities and arrhythmia onset, from the subcellular to organ level.
Sudden cardiac death, primarily caused by ventricular arrhythmias, is a major public health problem – it is one of the leading causes of mortality, resulting in more than 350,000 annual deaths in the United States alone. Our group’s efforts are focused on improving our understanding of, and therapies for, cardiac arrhythmias. We primarily investigate biophysical mechanisms of electrophysiological instabilities and arrhythmia onset, from the subcellular to organ level.Krogh-Madsen lab
Research Summary | Publications  Cardiac electrophysiology, control of cardiac arrhythmias, mechanisms of arrhythmogenesis, nonlinear dynamics and bifurcations, computational biology.
Cardiac electrophysiology, control of cardiac arrhythmias, mechanisms of arrhythmogenesis, nonlinear dynamics and bifurcations, computational biology.
Nimigean lab
Research Summary | Publications
 Research in our laboratory is geared toward understanding how ion channel protein structure and mechanism interrelate at the molecular level to allow channels to elaborate various biological properties. We use a combination of molecular, biochemical and electrophysiological approaches to evaluate in a complete fashion fundamental channel properties within the context of emerging structural data.
Research in our laboratory is geared toward understanding how ion channel protein structure and mechanism interrelate at the molecular level to allow channels to elaborate various biological properties. We use a combination of molecular, biochemical and electrophysiological approaches to evaluate in a complete fashion fundamental channel properties within the context of emerging structural data.Computational Genomics
Betel lab
Research Summary | Publications
 Our research interest is the development of computational genomic tools for the study of human diseases and cellular development. We focus on detailed analyses of genomic and epigenomic data generated by high-throughput sequencing to address specific questions related to disease progression, treatment response, stem cell differentiation and neurological processes.
Our research interest is the development of computational genomic tools for the study of human diseases and cellular development. We focus on detailed analyses of genomic and epigenomic data generated by high-throughput sequencing to address specific questions related to disease progression, treatment response, stem cell differentiation and neurological processes.Campagne lab
Research Summary | Publications
 Our laboratory develops computational methods and tools to facilitate new biological discoveries. We apply a variety of computational techniques, including data mining, visualization, information and knowledge management, machine learning and statistics to contribute to significant biomedical problems.
Our laboratory develops computational methods and tools to facilitate new biological discoveries. We apply a variety of computational techniques, including data mining, visualization, information and knowledge management, machine learning and statistics to contribute to significant biomedical problems.Mason lab
Research Summary | Publications
 We use functional genomics and high-throughput methods in three main areas: neurogenetics, tumor evolution, and synthetic biology. We leverage clinical cases which can reveal the most about the human genome: the most peculiar cases in human genetics, the most aggressive cancers, and the most striking brain malformations. Toward this end, we aim to use and improve technological methods in molecular genetics, so we can more accurately map the molecular networks of the genome and their changes over time.
We use functional genomics and high-throughput methods in three main areas: neurogenetics, tumor evolution, and synthetic biology. We leverage clinical cases which can reveal the most about the human genome: the most peculiar cases in human genetics, the most aggressive cancers, and the most striking brain malformations. Toward this end, we aim to use and improve technological methods in molecular genetics, so we can more accurately map the molecular networks of the genome and their changes over time.Hajirasouliha lab
Research Summary | Publications
 I am passionate about applications of computational and mathematical methods to molecular biology. My training is in computer science and I have been focused on computational genomics (including cancer genomics) and biomolecular sequence analysis in recent years. My research plans include characterizing cancer genomes and evolution. I have also experience in analyzing large-scale genomic data sets, and developed several methods for detecting structural variations in sequenced genomes. My research is highly collaborative and I worked with genome scientists, cancer biologists and clinicians on many recent projects. You can see the highlight of those collaborations in the Publications Section.
I am passionate about applications of computational and mathematical methods to molecular biology. My training is in computer science and I have been focused on computational genomics (including cancer genomics) and biomolecular sequence analysis in recent years. My research plans include characterizing cancer genomes and evolution. I have also experience in analyzing large-scale genomic data sets, and developed several methods for detecting structural variations in sequenced genomes. My research is highly collaborative and I worked with genome scientists, cancer biologists and clinicians on many recent projects. You can see the highlight of those collaborations in the Publications Section.Computational Cancer Genomics
Elemento lab
Research Summary | Publications
 We combine Big Data analytics with experimentation to develop entirely new ways to help prevent, diagnose, understand, treat and ultimately cure cancer. Our research involves routine use of ultrafast genome and DNA sequencing, proteomics, high-performance computing, mathematical modeling, and machine learning.
We combine Big Data analytics with experimentation to develop entirely new ways to help prevent, diagnose, understand, treat and ultimately cure cancer. Our research involves routine use of ultrafast genome and DNA sequencing, proteomics, high-performance computing, mathematical modeling, and machine learning.Hassane lab
Research Summary | Publications
 We are currently working in collaboration with other investigators in the Leukemia Program at Weill Cornell Medical College to develop a more detailed molecular understanding of LSCs. The ultimate goal is to develop therapeutic targets and state-of-the-art tests that are likely to positively impact patient care in the near term.
We are currently working in collaboration with other investigators in the Leukemia Program at Weill Cornell Medical College to develop a more detailed molecular understanding of LSCs. The ultimate goal is to develop therapeutic targets and state-of-the-art tests that are likely to positively impact patient care in the near term.Khurana lab
Research Summary | Publications
 My research focuses on the development of integrative computational models to understand the relationship between genomic sequence variation and disease. Functional impact of sequence variants in non-protein-coding regions of the genome is especially less-well-understood. We have developed an approach, FunSeq, to prioritize noncoding variants.
My research focuses on the development of integrative computational models to understand the relationship between genomic sequence variation and disease. Functional impact of sequence variants in non-protein-coding regions of the genome is especially less-well-understood. We have developed an approach, FunSeq, to prioritize noncoding variants.Sboner lab
Computational Molecular and Cell Physiology
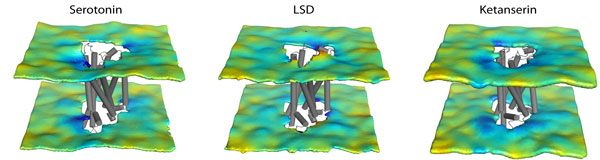
Shi lab
Research Summary | Publications
 Cell membrane separates the cellular machinery from external fluids. Membrane proteins (MP) initiate intracellular signaling pathways, control the flow of energy and materials in and out of the cell, and thereby account for more than 30% of proteome and 40% of drug targets. Research interests in the lab are focused on identifying common and specific structural basis of MP functions to advance the mechanistic understanding of key cellular processes, from the disparate yet intertwined perspectives of Structure-Dynamic-Function Relationships and Molecular Recognition.
Cell membrane separates the cellular machinery from external fluids. Membrane proteins (MP) initiate intracellular signaling pathways, control the flow of energy and materials in and out of the cell, and thereby account for more than 30% of proteome and 40% of drug targets. Research interests in the lab are focused on identifying common and specific structural basis of MP functions to advance the mechanistic understanding of key cellular processes, from the disparate yet intertwined perspectives of Structure-Dynamic-Function Relationships and Molecular Recognition.Weinstein lab
Research Summary | Publications
 Structural, dynamic and electronic determinants of biological processes underlying physiological functions are addressed in the lab through the development and application of methods in theoretical and computational biophysics. Much of the computational and modeling efforts offer improved methods and new structural insights related to function. The approaches include theoretical determinations of molecular structure and properties, computational simulations and bioinformatics.
Structural, dynamic and electronic determinants of biological processes underlying physiological functions are addressed in the lab through the development and application of methods in theoretical and computational biophysics. Much of the computational and modeling efforts offer improved methods and new structural insights related to function. The approaches include theoretical determinations of molecular structure and properties, computational simulations and bioinformatics.Statistical Genetics
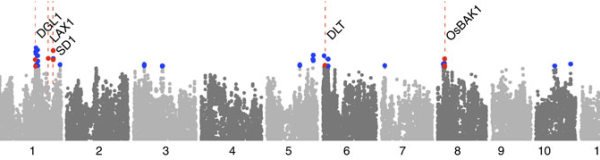
Mezey lab
Research Summary | Publications
 We are a computational genomics group focused on understanding the genetics, development, and evolution of complex phenotypes and disease. Our research is primarily computational, involving the development and application of techniques that fall within the disciplines of computational statistics and machine learning, although we also complement these efforts with our own experimental research.
We are a computational genomics group focused on understanding the genetics, development, and evolution of complex phenotypes and disease. Our research is primarily computational, involving the development and application of techniques that fall within the disciplines of computational statistics and machine learning, although we also complement these efforts with our own experimental research.
